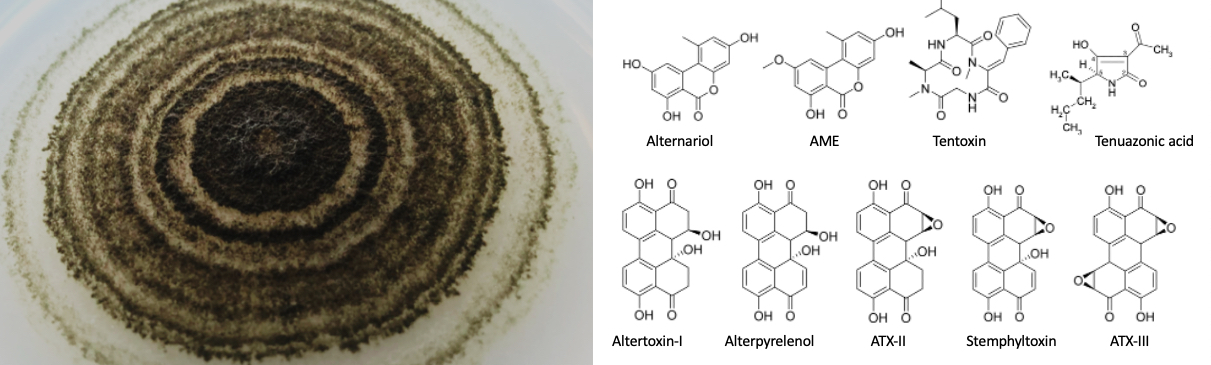
Alternaria alternata produces a large variety of mycotoxins. More than 70 different molecules were described. Many secondary metabolites are derivatives of polyketides and hence the number of polyketide synthases encoded in the genome is an indicator for the number of substances which can be produced. However, the genome of A. alternata harbors only 10 polyketide synthase encoding genes. The alternariol gene cluster is a good example how with different additional enzymes encoded in the gene cluster different variants can be produced. In the case of the alternariol gene cluster, alternariol, alternariol methyl ether (AME), hydroxy-AME, alternusin, and alternuene are produced from the same precursor.
The goal is to assign a function to all ten gene clusters. This was very difficult in the past, because genome modifications were quite laborious, because genes could not easyily deleted by homologous recombination. Therefore, we established the CRISPR/Cas9 system in this fungus. Since then, we are able to generate routinely gene deletion strains.
Analysis of the alternariol biosynthetic gene cluster
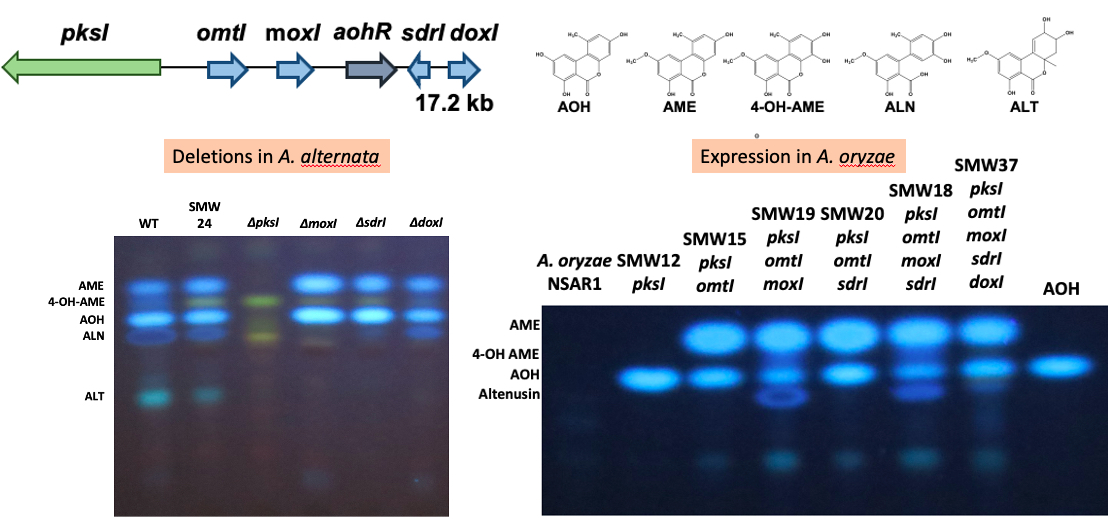
This figure illustrates our strategy to design a function to gene clusters in A. alternata with the example of the alternariol gene cluster. We delete individual genes and analyze the metabolites in extracts of A. alternata by thin-layer chromatography (left) or by HPLC. Some bands are missing in corresponding deletions strains. On the other hand, we are able to express several genes at the same time in Aspergillus oryzae (right panel). The transgenic strains are then analyzed for the production of new metabolites. For more details see the paper of Wenderoth et al., 2019.
